Introduction
Aromatic amines are toxic and mutagenic compounds formed during the reductive cleavage of nitro-aromatics and azo dyes [1]. Some of the compounds are produced by incomplete combustion of fossil fuels and others are flushed out from the industries of dyes, drugs, pesticides, and explosives. Nitro-aromatics can act as inhibitors of nitrification; as the di-nitro-aniline belongs to herbicide group widely used against weeds. Many microbial strains are capable of transforming nitro aromatic compounds by various ways like aerobic or anaerobic [2–4]. Generally anaerobic bacteria reduce the nitro group via nitroso and hydroxylamino intermediates to the respected amines. The anaerobic bacteria and fungi are generally mentioned to transform nitro-aromatics. Some of the bacteria reduce the aromatic ring having di-nitro and tri-nitro group by the addition of a hydride ion and form ahydride-meisenheimer complex, which subsequently rearomatizes and eliminates nitrite [5,6]. Monooxygenase enzyme adds a single oxygen atom to the nitro-aromatic molecule and eliminates the nitro group from nitro-phenols. The dioxygenase enzymes insert two hydroxyl groups into the aromatic ring and participates spontaneously in the elimination of the nitro group. The reduction of the nitro group forming hydroxylamine is supposed to be the initial step of degradation of such compounds [7]. This hydroxylamine undergoes enzyme catalyzed rearrangements and further hydroxylated product leads to ring-fission. Various reports are elaborating the detoxification of such contaminants in neutral condition. Few of the reports are available in alkaline condition [8–14]. Although these researchers have been worked using alkaliphiles; they studied the bio-degradation of hydrocarbons, phenolics, cyanides, thio-cyanates, lignin, azo-dye etc. Earlier to it Olga Maltasalva and others studied the biodegradation of 2, 4 dichlorophenoxy acetic acid by using three haloalkaliphilic isolates on mineral medium. It was observed that the microbial strain employed was more active and it degraded 2, 4D (3,000 mg/L) within 3 days by ortho cleavage pathway at pH 8.4 to 9.4 with salinity 0.6 to 1.0 M. in aerobic condition [15]. In one another report it was noticed that halophilic marinobacter sp able to metabolize 1, 3-diphenylurea at 35 °C and 0.51 M salt concentration [16]. The bacterial strain Dietzia psychralcaliphila was observed growing on n-alkane as carbon source [17]. A species of halomonas degraded several organic acids and predicted that could be useful for the decontamination of such polluted sites [18]. Similarly, in one another study a halophilic strain of fungus penicillium chrysogenum grown at 5–6% salinity and degraded 300 mg/L phenol without accumulation of any intermediates [19].
Recently researchers have been reported that azo reductase from alkaliphilic bacteria bacillus badius shown nitroreductase activity along with azoreductase. In this report the researchers were studied 3-nitrobenzoic acid, 4-nitro toluene, 3- nitro toluene and 1-chloro 2 nitro-toluene enzymatically [20]. However this present study employing pseudomonas DL17 is apart from their work. Here an attempt was made to search the pathway of p-nitroaniline degradation in alkaline habitat using live bacterial cells of alkaliphilic strain pseudomonas DL17 isolated from Lonar Lake, Buldana (MS) India.
Significance of this study
Biodegradation is an important environmental process to remove contaminants and clean up the environment. Study of biotransformation gives idea about the enzymes involved in it as well as the novel bio-transformed compounds which has very much importance itself. Detoxification and decontamination are very important processes for healthy ecosystem. During this process one can isolate the important chemical compounds which are supposed to be wasted. Polluted water invites various biogenic or chemical born diseases to human beings. The nitro-aromatic compounds can cause cancer and kidney damage. Microbes are good source of remedy to decontaminate such pollutants. This study is significant to bioremediation of nitro-aromatics contaminated sites and saving the lives from its exposure. It could make possible to isolate few chemicals such as the metabolites generated and can be reused as raw material. It also gives a new source of the bio-transformation enzymes as well as other biozymes for making harsh chemical reactions easy.
Methodology
Chemicals
Organic and inorganic chemicals were demanded from SRL, Mumbai, and Yeast extract peptone and other bacterial media were purchased by Hi media Mumbai.
Isolation of alkaliphiles
Soda lakes are the most stable and productive naturally occurring alkaline environments in the world with pH higher than 10 and occasionally reaching pH12.0. The soil collected in the form of sediment from Lonar Lake Buldana, (MS) India; and dissolved in sterile sodium bicarbonate buffer pH 9.0 aseptically. The isolation of pure strains of microorganisms was done by serial dilution and pour plate technique. During serial dilution of soil sample10 small sterile testtubes were taken and labeled from 1 to 10. Then 4.5 mL sterile physiological buffered saline was added to each test tube. By the pipette 0.5 mL of the original suspension transferred into test tube no.1. This bacterial suspension was mixed thoroughly before proceeding to the next step. From first test-tube 0.5 mL of the diluted bacterial suspension withdrawn by clean sterile pipette and added to second test tube. It was continued till the dilution in 10th test tube. In test tube no.1 the bacteria diluted were 10 fold, as 1:10 or 1 × 10−1. Similarly the dilution of the last test-tube was 1 × 10−10 carried. Using the pipette, 0.5 mL aliquots from each test tube was poured and spreaded by sterile spreader to get isolated colonies on respected petriplate. The petriplates were incubated at 37°C for 24 h. Four isolated colonies were observed on alkaline agar plate among which two were slight bigger than others [21,22]. One of them was faint yellow colored. Both were tried to search for its nitro-aromatic degradation potential and the yellow-colored colonies were proceeded for nitro-aromatic amine degradation study due to its immediate response to p-nitroaniline showing resistance in alkaline agar and broth. A single isolated colony of it was further used for sub culturing on alkaline agar slants for further studies.
Media content for experiments
The media composition were similar to earlier work [23], containing yeast extract, peptone, NaCl-5 g/L respectively and the micronutrient’s in mg/L were KH2PO4 −170, Na2HPO4 −290, (NH4)2SO4 −100, MgSO4, MgO-0.1, FeSO4 −0.05, CaCO3 0.20, ZnSO4 −0.08, CuSO4 −0.016, CaSO4 −0.016, Boric acid 0.06, and pH 9.0. The pH was adjusted by sodium bicarbonate (0.1 M or 1 M) solution. The media was sterilized by autoclaving at 121 °C for 15 pounds at 20 minutes. The solid media was prepared in same way by adding 2% agar for petriplates and agar slants.
Identification of the microorganism
The DNA from the bacterial cells was extracted using a typical DNAzol method by Saitou, N .and Nei M. 1987. Approximately 9 colonies of pure culture from petriplate were suspended in 1× TE buffer and washed twice in 500 μL TE by vortex mix followed by centrifuge at 10,000 × g for 10 minutes at room temperature. Bacterial pellets then suspended to 100 μL protease K (at the final concentration of 325 μg/mL) at 65 °C for 1 h and 30 minutes. Lysis was done by DNAzol treatment as 500 μL for bacterial pellets with proteinase K mixture at 65 °C for 5 minutes. Upper DNAzol phase was carefully separated onto 500 μL of absolute chilled ethanol resulting in bacterial DNA precipitation followed by two washing of 70% cold ethanol. DNA pellets were collected at 10,000 g for 10 minutes at room temperature and solubilized in 100 μL of TE buffer. For each 100 PCR reaction 500 ng templates of DNA were used. 16S rRNA method was employed for identification of bacteria at molecular level which showed maximum similarity with pseudomonas after blast [24]. The strain was submitted to NCBI with its sequence JN595813.1 and named as pseudomonas DL17.
Bio-degradation study
Nine 500 mL conical flasks with 200 mL alkaline broth media were used for biodegradation study using 24 h grown bacterial culture (1%) as an inoculum. This was again allowed to grow for 24 h in shaking incubator at 37 °C, 110 rpm. At the end of 24 h (500 mg/L) p-Nitro-aniline was added in each flask aseptically. P-nitro-aniline degradation was monitored by UV-vis spectrometry by 6 h interval after spinning it using cold centrifuge at 10,000 g. Residual p-nitro-aniline was estimated spectrophotometrically by piepetting an aliquot of supernatant and after proper dilution at absorbance at 350 nm and by plotting standard graph [25].
Purification and characterization of residual remains
Remaining supernatant by using the aliquots further used for the extraction of metabolites generated from p- nitro-aniline during biotransformation. The solvent extraction was generally carried in organic solvents like ethyl acetate or DCM. The residue obtained by solvent extraction and rotary evaporator further purified by preparative TLC or column chromatography. The structural determinations of the metabolites were carried out by spectroscopic analysis like FTIR, 1H-NMR and GCMS study.
Preparation of cytosolic fraction
The cell mass was obtained by induction with experimental concentration of p-Nitro-aniline for 24 h and it was washed thrice by phosphate buffer pH 8.0. The washed cell was ruptured by sonication in phosphate buffer of same pH and homogenized by vortex mixture. The cellular debries were removed by cold centrifuge at 15,000 g. This supernatant was used for enzymatic study.
Enzymatic study
Nitroreductase activity experiments were performed by slight modification of Christopher Bryant method [26]. The assay mixture contained 45 μM p-nitroaniline and 0.30 mM NADPH making 1.0 mL final volume using 50 mM buffer PH (7.0). One unit of enzyme activity was analyzed as quantity of enzyme required to oxidize 1 μmole of NADPH per minute at 25 °C and was measured by linear decrease in absorbance at 340 nm. The acetanilide hydroxylase activity was estimated using 1.25 μL 50 mM Tris buffer pH (7.4) added with 1 mL of 0.03M NaCl kept at 0 °C for 10 min followed by 1 mL 0.03M R nitro salt in 0.5N NH4OH. Further it is added with 1 mL 8 mM acetanilide and 0.2 mg/LNADPH making total volume 5 mL. After attaining redish color it was measured at 484 nm and compared with standard by graph as per Weisberg and Goodall [27]. For the determination of superoxide dismutase activity total reaction mixture 1 mL contained 960 μL of 0.05 M carbonate buffer pH (10.2) 20 mM epinephrine in 0.1 m MEDTA 40 μL. The androchrome formed by auto-oxidation was monitored at 480 nm with 40 seconds intervals for 5 minutes at room temperature. Similarly the inhibition of auto-oxidation of substrate epinephrine measured by using same ingredients with addition of 20 μL of cytosolic fraction as an enzyme source [28]. Catechol-1, 2-dioxygenaseactivityanalyzed spectrophotometrically knowing the formation of cis-cis muconic acid at 260-nm (ɛ-260-16,800M−1cm−1). The reaction mixture in 1 mL quartz cuvettes had 980 μL Tris buffer pH (7.5)and 10 μL 20mM catechol with 10 μL enzyme source[29]. Estimation of Catechol 2,3 dioxygenase was carried by using 300 μL cytosolic fraction mixed with 700 μL catechol 10 mM in 50 mM phosphate buffer pH (7.5) using molar coefficient (ɛ-3300 M−1cm−1) at 375 nm wavelength [30]. The activities of catalase and peroxidase were determined by Zeng [31]. Catalase neutralizes the H2O2 generated in bacteria. It differentiates between aerobic and obligate anaerobic bacteria. For the determination of catalase the reaction mixture contain 100 μL enzyme source 1 mL 50 mM KH2PO4 buffer pH (7.0) and 1 mL 15 mM H2O2. The total volume made 4 mL with distilled water and readings were taken at 420 nm. Unit activity was measured as the amount of enzyme required to transform 1 μm of H2O2 to H2O per min. The molar constant used for H2O2 was (ɛ-43.6 /m−1 cm−1). Peroxidase activity was measured at 470 nm at 32 °C. The 4 mL volume contain 100 μL enzyme source, 50 mM Na2HPO4-citrate buffer pH 6.0 and 10 mM H2O2 with 10 mM gaucol [32].
Results
UV-vis spectroscopic study
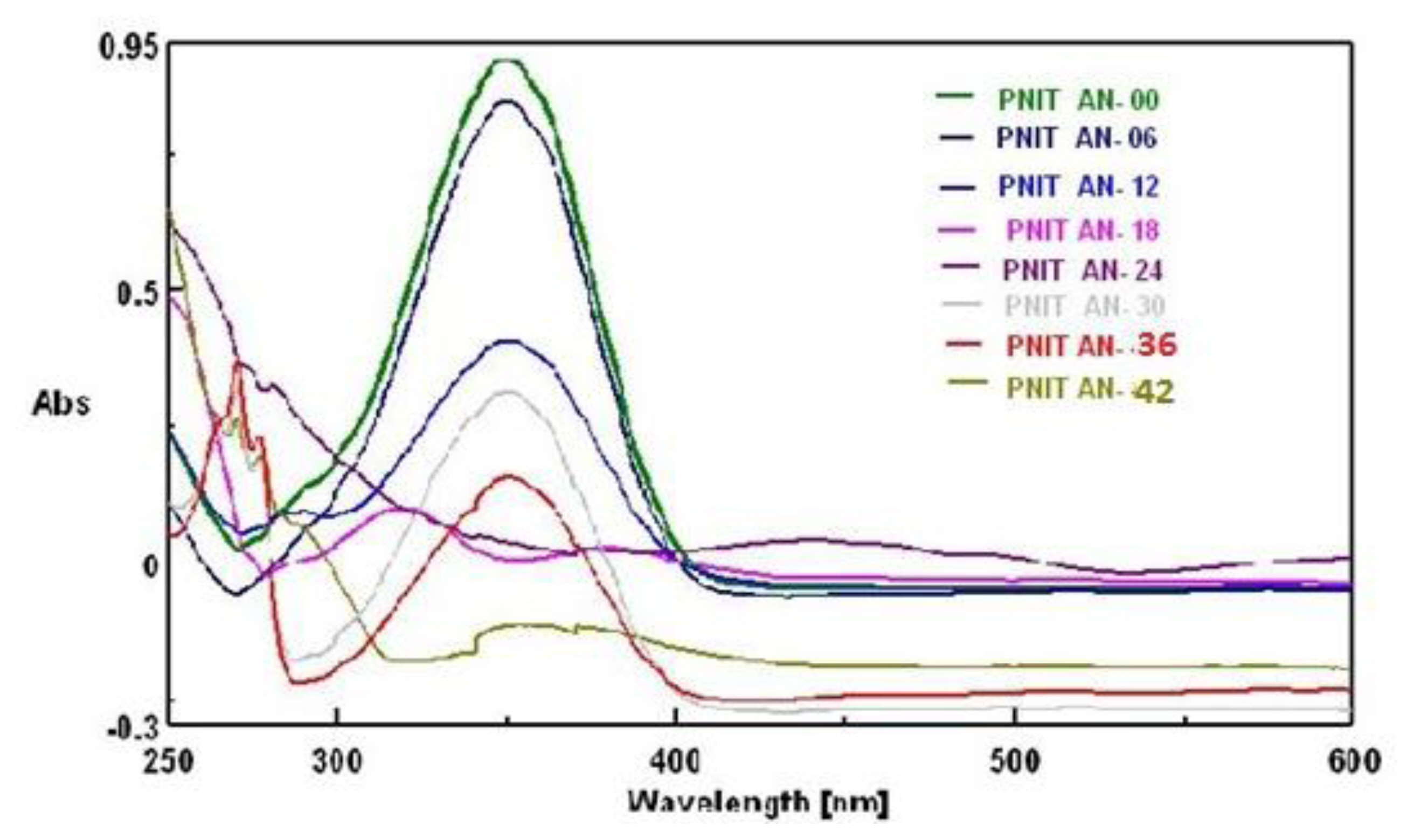
It has been observed that the initial spectrum of p-nitro-aniline at 0 h. having high absorbance at 350 nm used as a standard shown in Figure 1 reduced after 6 h suggested that the experimental strain might have consumed certain p-nitro-aniline. Even though at 12 h the reduction in absorbance was noticed little less. Minute changes might have occurred in the moiety; however one of the spectra at 18 h showed major change in absorption wave length by its peak at 324 nm could be the spectrum of new metabolite formed. It was further confirmed by HNMR, FTIR and GCMS as p-phenylenediamine. It was also noticed that this metabolite p-phenylenediamine showed its appearance in between 18 to 30 h. By TLC its ‘rf’ value found 0.73 in ethyl acetate: n-butanol: water (6:2.5:1.5) phase and confirmed with standard. The 42 h spectrum showed least absorption and its most of the part found below the ‘0’ base line therefore no further spectrum collected.
It is known that microbes can use nitro substituted aromatic compound as carbon and nitrogen source [33]. The strain pseudomonas DL17 is an indigenous strain tolerated (shown resistance) to higher concentration of p-nitro-aniline on adaptation to one month. It showed slowly decolorization of p-nitro aniline amended broth within 24h; while no change in control flask (abiotic control-heat killed bacterial cells of experimental organism) was observed. Therefore the experimental organism was selected for biodegradation study. The adaptation was initiated with (50 mg/L) of p-nitro aniline augmentation in 500 mL Erlenmeyer flasks with 200 mL alkaline broth at pH 9.0. Although the experimental chemical is toxic; it shown visible growth in the broth media. Thus the augmentation of p-nitroaniline further increased till (500 mg/L). After knowing the optimum temperature 32 °C and pH 9.0 for pseudomonas DL17 experiments were designed to know the degradation and biotransformation.
Biodegradation study
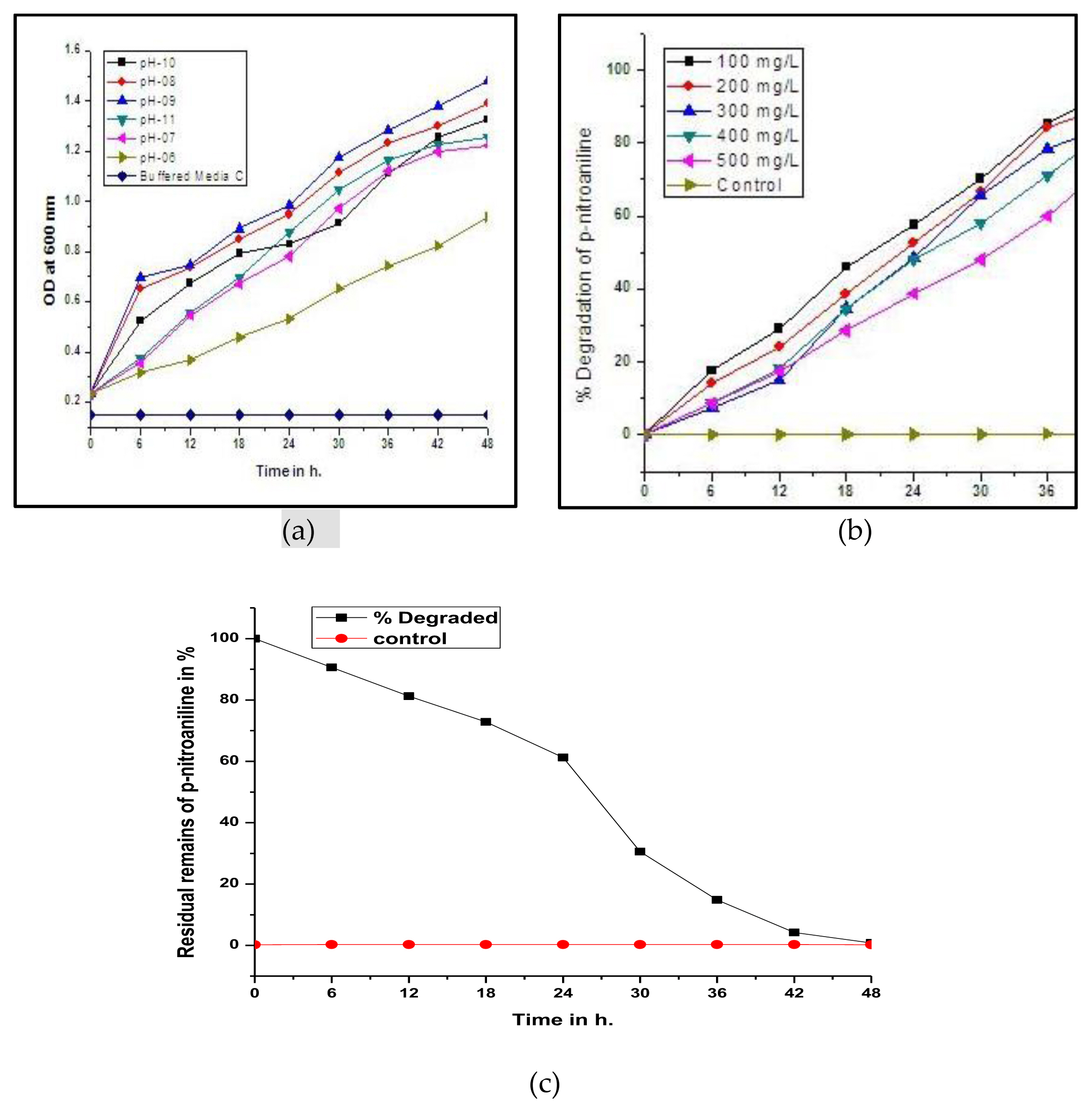
Extremophiles are the microorganisms which grow in environments of extreme temperature such as (−2 to 15 °C, 60–110 °C), ionic strength (2–5 M NaCl) or pH (<4, >9) or high pressure [34]. As the bacterial strain pseudomonas DL17 was isolated from alkaline habitat Lonar Lake, Buldana (MS), India. It has preferred pH 9.0 for growth [35] shown in Figure 2 (a). The results obtained showed that Figure 1 and 2 (b) it has consumed p-nitro aniline as an additional carbon and nitrogen source. Aliquots from broth (working and control flasks) were collected after centrifugation and used to confirm residual p-nitro-aniline concentration parallel with change in UV-vis spectral study. Marginal difference in % degradation of experimental compound was observed at pH 9.0 on varying p-nitro aniline (100–500 mg/L) concentration in working flasks. However no change in the (heat killed bacterial cells) abiotic control flask was noticed. Further adaptation to 15 days pseudomonas DL17 degraded the p-nitroaniline (500 mg/L) within 48 h. Thus the experimental bacterial strain seems useful for bioremediation purpose. Particularly at the pH 9.0 it showed 100% degradation and it was noticed in Figure 2 (c).
The higher concentration of p-nitroaniline (500 mg/L or more than it) might found toxic due to formation of catechol or hydroquinone or bezoquinone like molecules in excess generating oxidative stress on the cell and even affecting DNA [36]. Figure 5 showed that bezoquinone have been generated during the catabolism. Metabolite isolated by column chromatography showing similar spectrum with p–bezoquinone.
The source of bezoquinone or semi-bezoquinone formed might have come from hydroquinone or more hydroxyl aromatic compounds generated with catechol. Figure 3 showed formation of p-bezoquinone at 36 h as a metabolite and confirmed by higher spectrophotometric methods. This might be due to saturation of phenolic compounds such as catechol in metabolic process exerting oxidative stress on cell and for diverting the toxicity generated due to higher catechol appeared during metabolic process. It is supported by the induction activity of super oxide dismutase, catalase and peroxidase shown in Figure 7. Higher concentration of phenolic compounds generated such as catechol might have caused metabolic stress and lead to activation of ortho and meta cleavage pathways. The FTIR, 1HNMR and GCMS level study confirmed various bio-transformed metabolites such as p-phenylenediamine, acetanilide, acetaminophen (paracetamol), catechol, cis-cis muconic acid etc. by which the p-nitroaniline mineralization mechanism was deduced.
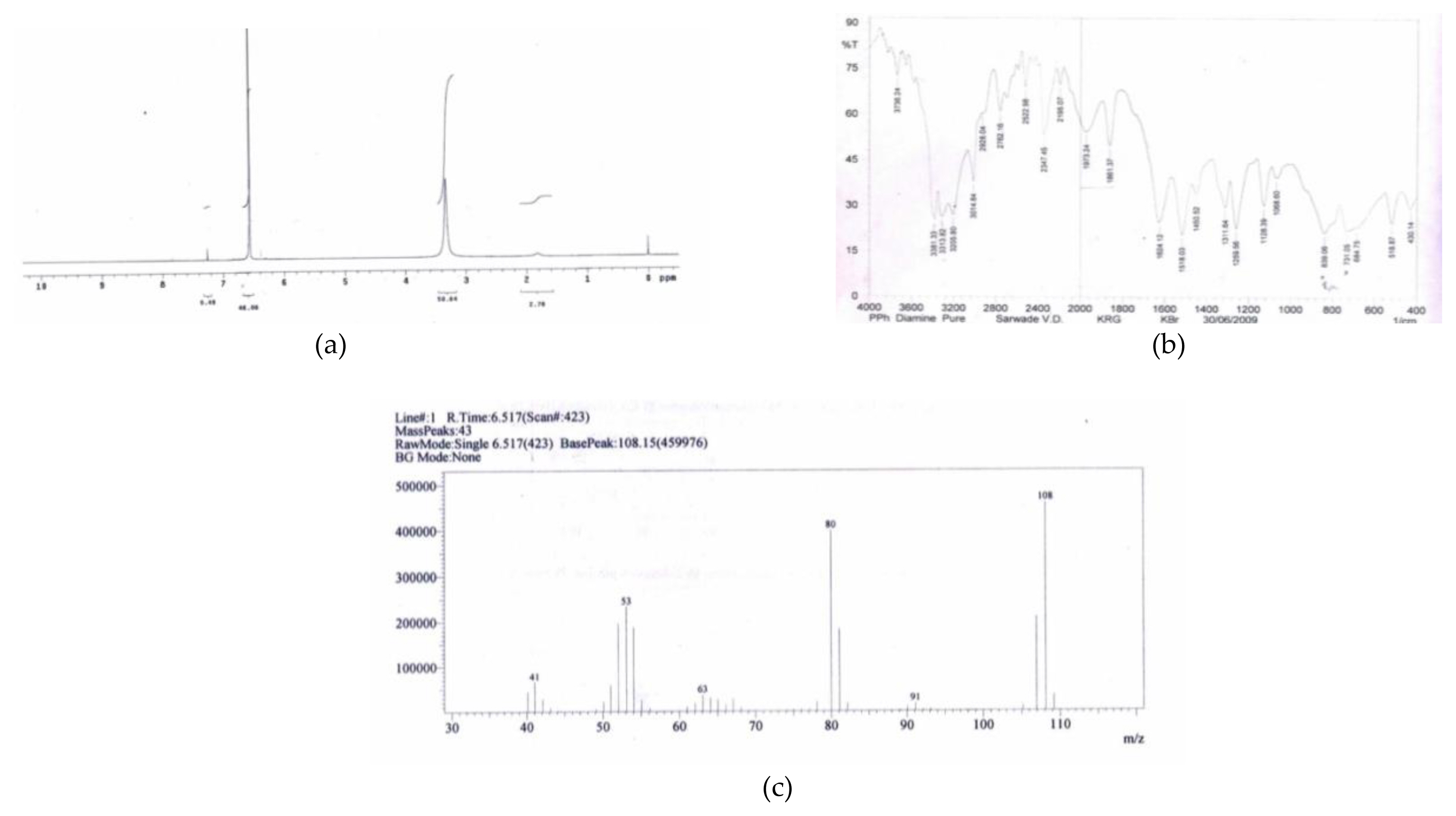
p-phenylenediamine (a) 1HNMR, (b) FTIR and (c) mass spectral data obtained after analysis of metabolite
The Figure 4 (a) showed ‘δ’ −3.33 (S-4H), 6.65(S-4H) due to similar position of hydrogen present on nitrogen of p-phenylenediamine in between ‘δ’ 3 to 4 ppm and in the area of aromatic ring ‘δ’ 6 to 7 ppm. It showed two prominent singlet peaks in this spectrum. Figure 4 (b) FTIR spectrum shows stretches Ar-NH2-,3311, and for NH2at Ar-1518 confirmed the functional groups. Figure 4 (c) showed the molecular ion peak at 108 and the fragmentation pattern 108,91,80,63,53,41 etc. satisfied it as p-phenylenediamine in mass spectrometry.
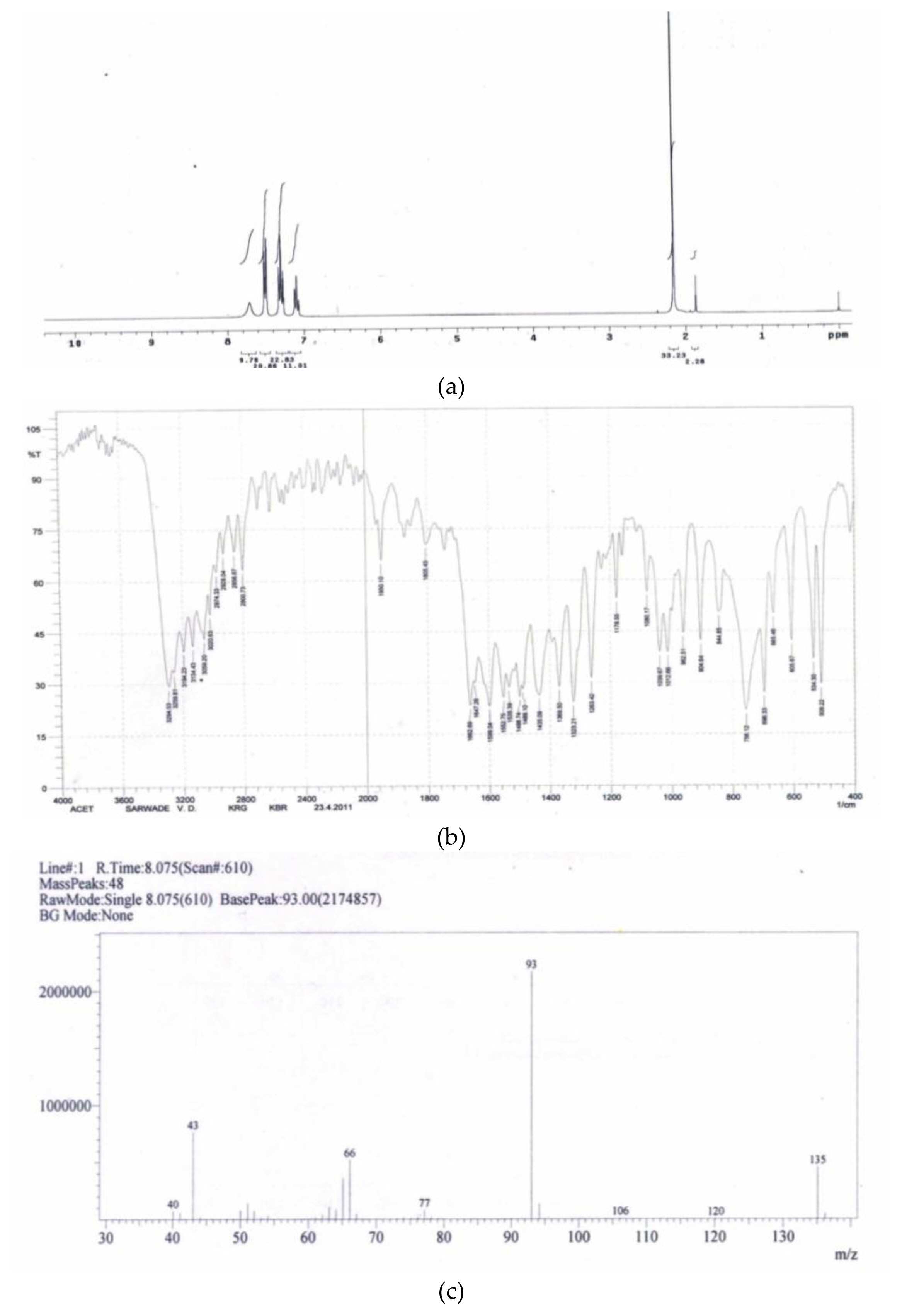
Acetanilide: (a) 1HNMR (b) FTIR (c) mass spectral data
The Figure 5 (a) showed pattern of δ values as δ-2.151, (S-3H), δ-7.5, (m-2H), confirming it as acetanilide. Figure 5 (b) showed stretches at AR-C=O-1662, AR-1498-1535, AR-NH-3294 and confirmed -NHCOCH3 group stretches in aromatic region confirming it as acetanilide. Figure 5 (c) fragmentation pattern 135 as molecular ion peak, 93 as base peak and rest are 77, 66, 43, as fragments confirms it as acetanilide similarly other metabolites were confirmed and provided as supplementary data.
Mechanism of mineralization
The Figure 6 shows the probable pathway of mineralization of p-nitroaniline by pseudomonas DL17. The metabolites isolated and confirmed by sophisticated techniques like UV-Vis spctrophotometry as acetanilide, aniline, p-Phenylediamine, catechol etc. However the bracket shown metabolite were only expected to be formed and not trapped during experiment. Acetaminophen or p-hydroxyacetanilide is also known as paracetamol. The Table 1 shows data analyzed by H-NMR, FTIR and mass spectrometry. These compounds are important to industries as a raw material. Agmenellum quadruplicatum strain PR-6 and oscillatoria sp strain JCM grown photoautotrophically in the presence of aniline metabolized the aromatic amine to formanilide, acetanilide and p-aminophenol [37]. In one another study on administration of small dose of aniline to male volunteers in human being resulted n-acetyl-4-aminophenol as the predominant urinary metabolite after LCMS analysis [38]. Rhodococcus erythropolis AN-13 when grown on aniline it was found that cis-cis-muconic acid and β-ketoadipic acid as a metabolite which were confirmed by thin-layer chromatography [39]. Epidermal cells and hepatocytes biotransformed PPD (p-phenylenediamine to n-mono-(MAPPD) monoacetylated and N, N’-di-acetylated (DAPPD) phenylenediamine derivatives [40]. One of the metabolites found in azo dye degradation was p-Phenylenediamine during its mineralization [41]. The nitroreductase enzyme family is able to metabolize nitro-aromatics using the reducing power of flavin based NADH/NADPH or NADPH independently [42]. Similar strategy might have been used by the experimental strain pseudomonas DL17. Nitroreductase have biotechnological application in bio-catalysis, clinically as a chemotherapeutic agent for tumor treatment, ablation of specific cells [43]. An aryl amine N-acetyltransferase (NAT) which is responsible for the N-acetylation of p-phenylenediamine [44]. Carboxylases are the enzymes important in the biosphere, having role in the global carbon cycle for the fixation of inorganic carbon as CO2. It is also having important role in carbon assimilation in biosynthetic pathways and in redox-balancing functions. Carboxylases are the enzymes that catalyze the incorporation of a CO2 molecule into an organic substrate and increase the polarity of contaminant molecules making easy for bio-transformation [45]. Catechol 1, 2 dioxygenase and catechol 2,3 dioxygenase are important ring cleavage enzymes that helps in mineralization of aromatic compounds [46,47].
Enzymatic study of pseudomonas DL17
Microbial enzymes are the biocatalyst which performs reactions in environmental friendly way as well as it avoids the harsh chemical processes. Bacterial enzymes have proved their utility in industries such as food, leather; textiles, animal feed, and in bioremediations [48]. A popular analgesic-antipyretic acetaminophen was synthesized from aniline by successful protoplast fusion between S. lividans and S. globisporus as well as between S. rimosus and S. aureofaciens. Thus it was known that the acetanilide hydroxylase is having commercial importance [49]. Many industries are making efforts to move away from the use of harsh chemicals and tedious or risky reactions; as a result there is increasing biotechnological demand for enzymes which are stable at extreme condition.
The Figure 7 clearly indicates the induction of nitroreductase, acetanilide hydroxylase, peroxidase, catalase catechol 1, 2 dioxygenase, catechol 2, 3 dioxygenase and super oxide dismutase than standard. It indicates that the experimental strain pseudomonas DL17 is actively participated in biotransformation process. Plants and microorganisms produce a wide range of oxidative enzymes e.g. peroxidase, polyphenoloxidase, and tyrosinase other than CYP P450s. The cytotoxicity of nitro aromatics in spleenocytes increases with an increase in their single-electron reduction potential. It points to the prevailing mechanism of the oxidative stress-type cytotoxicity. At higher concentration of p-nitroaniline might cause to generate oxidative stress by which superoxide dismutase have been induced [50]. Monofunctional catalases oxidizes H2O2 directly to molecular oxygen which prevent the damage of cell from free hydroxyl radicals [51].
H2O2 is powerful oxidizing agent used in industries for paper and cloths for belaching. As it is concerning with cell it is important to prevent such damage by catalases, superoxide dismutases etc. The (SODs) are a group of metalloenzymes found in all kingdoms of life. SODs form the front line defense against reactive oxygen species mediated injury [52]. Peroxidase is a heme containing enzyme which catalyzes the oxidation of a wide variety of organic and inorganic substrates using hydrogen peroxide as the electron acceptor. These are involved in decomposition of pollutants such as paper and pulp industries as well as various dyes as an effluent. Mostly these are the enzymes used in decolorization of synthetic dyes, pigments, chlorinated compounds and phenolics [53].
Discussion
Biotransformation is part of biodegradation process. It may involve several successive enzymatic reactions (Figure 6) with the chemical compounds as substrate and products. As it is known that neutrophiles could not tolerate alkaline condition so far and hence to study the mechanism of detoxification in alkaline condition an indigenous alkaliphile pseudomonas DL17 was employed. Alkaliphiles thrive at pH of 9.0 and above which are represented by archaea prokaryotes, eukaryotes and could be applied in the biodegradation and (or) biotransformation of various toxic industrial pollutants as well as brackish wastewater treatment. As the cell wall of these bacteria plays significant role in maintaining the intracellular pH homeostatically or symports and anitiports with additional Na+/K+ pumps [54]. Having the advantage of such tolerance to alkalinity and salinity; the p-nitro aniline was found totally degraded by the experimental strain Figure 1 and Figure 2(c). Marginal difference in % degradation was found when various concentrations were used. In case of Figure 2(b) at (500 mg/L showed little less % degradation than others but on further adaptation for 15 days made possible to degrade it all within 48h (Figure 2(c)). It has preferred pH 9.0 for optimum growth Figure 2(a). Microbial consumption of nitro aromatics compounds was reported by some other researchers on succinate augmentation [55]. Microbes of the bovine rumen fluid catalyzed the reduction of nitro compounds as substrates and yielded the respective amines. This enzymatic process, using ruminal contents, was rarely reported in case of bio-reduction of nitro groups [56]. Degradation or detoxification pathways include oxidation, reduction, hydrolysis, and conjugation. Metabolic pathways and its diversity depend on the chemical structure of the contaminant, nature of the organism, environmental conditions, metabolic factors, and the regulating expression of these biochemical pathways [57]. Oxygenation was found the most frequent first step in the biotransformation of pesticides and other organic contaminants. A bacterial strain of stenotrophomonas HPC 135 found capable of growing on 4-nitroaniline as a source of carbon and energy similar to the experimental bacterial strain [58].
The nitro-aromatic compounds are being used worldwide as an explosives, pesticides, and as a precursors for the manufacture of many products, including dyes, pharmaceuticals, and plastics [59]. The nitro-aromatic compounds are well known toxins. Many much among them are found mutagenic and/or carcinogenic [60,61]. These compounds are uncouplers of cellular phosphorylation [62]. Many mono and di-nitro-aromatic molecules are readily degraded by aerobic bacteria through a variety of monooxygenase or dioxygenase based pathways. Anaerobic bacteria employ reductive pathways, often exploiting their low redox potential electron transport systems to get the things started [63]. No effect on growth of stenotrophomonas HPC on acetate augmentation observed but reduction of 4-nitro-aniline was noticed by earlier researcher. However candida guillermondii transformed the nitrobenzene into aniline [64]. The subsequent acetylation of amines leads to the corresponding acetanilide or amides. Non-polar nitro-aromatics have been considered resistant to attack by oxygenase due to the electron withdrawing effect of nitro group [65]. The nitro-aromatic amines and azo dyes are highly toxic to acetoclastic bacteria. However these carcinogenic contaminants were detoxified by some methanogenic consortia to respective aromatic amines which are several orders less toxic than earlier one [66]. A strain of pseudomonas P6 utilized p-nitro-aniline as a sole source of carbon and brought its mineralization by 8 days. However the experimental strain pseudomonas DL17 consumed it within 48 h. [67]. Other bacterial strain burkholdaria terrae KU 15 degraded 2-nitrobenzoate and produced 3, hydroxyl anthranilate as a metabolite [68]. Even the preferential reduction of nitro group in the ortho position due to resonance electron-donating groups such as NH2 or OH also reported for both chemical and enzymatic reduction [69]. Hydroxylamino aromatic compounds are converted to aminophenols or protocatechuate during the bacterial degradation of nitro-aromatic compounds [70].
The experimental strain pseudomonas DL17 degraded 100% p-nitroaniline (500 mg/L) within 48h. The first important step in the catabolism of nitro-reduction is followed by the NADPH dependent nitroreductase. The oxygen-insensitive and type I nitro-reductases are flavozymes which mediate the sequential transfer of two electrons from NADPH to the nitro group and produce nitroso compounds as well as hydroxylamine derivatives subsequently removing ammonium ions through Lyase-mediated reaction and leading to catechol pathway [71]. The mutase mediated reaction such as Bamberger-type rearrangement occurred in some cases to produce 2-aminophenols or benzoates resulting in release of ammonia suggesting another route of catabolism [72]. Thus by the results obtained by various observations it can be concluded that the experimental bacterial strain pseudomonas DL17 is actively involved in biodegradation process. Nitro-aromatic compounds which are normally considered to be toxic to microorganisms caused the induction of drug metabolizing enzymes such as cytochrome P-450, aminopyrine N-demethylase, acetanilide hydroxylase and glutathione S-transferase. These enzymes constitute the bacterial mixed function oxidase system. The p-phenylediamine generated after reduction by nitroreductase might have come across the aryl amine N-acetyl transferase and it immediately have been transformed into N-acetyl derivative. This was reported as xenobotic metabolizing enzymes involved in the detoxification of numerous aromatic chemicals [73]. 2,4-Diamino-6-nitrotoluene (2,4-DANT) was transformed by pseudomonas fluorescens to a novel metabolite, 4-N-acetylamino-2-amino-6-nitrotoluene and the other-one 2,6-diamino-4-nitrotoluene (2,6-DANT) which was observed persistent [74]. Similarly the acetanilide formed in this experiment might have transformed into acetaminophen or p-hydroxyl acetanilide by acetanilide hydroxylase [75]. Figure 1 showed several changes in p-nitro-aniline spectrum indicating biotransformation in the p-nitro aniline molecule and its consumption [76]. The catechol formed as part of central metabolite might have undergone ortho or meta cleavage pathways activating catechol 1,2-dioxygenase and or catechol 2,3-dioxygenase transforming it to cis-cis muconate by ortho cleavage pathway, while 2-hydroxymuconic acid semialdehyde by meta cleavage pathway leading to TCA cycle [77,78]. The experimental bacterial strain have showed tolerance to the p-nitroaniline as earlier stated suggesting the involvement of its resistance factors present on the main genome or on the plasmid. Similarly Figure 2(c) indicated the degradation of higher concentration of contaminant by the experimental strain. Almost 100% of (500 mg/L) p-nitroaniline was found exhausted within 48 h. The experimental strain pseudomonas DL17 showed the favorable pH range 8.0 to 9.0 for growth Figure 2(b). Figure 3 showed one of the metabolite bezoquinone appeared in the broth indicating higher concentration of catechol being produced in metabolic process and to divert the effect of oxidative stress generated it might have activated the hydroquinone pathway. This hydroquinone further might have transformed in bezoquinone. Induction of catechol 1, 2 dioxygenase and catechol 2,3 dioxygenase revealed the higher concentration of catechol in its metabolic process. In case of the metabolism of experimental strain pseudomonas DL17 one of the intermediates found was p-phenylenediamine indicated that the first step is reduction and as a key step similar to other oxygen insensitive nitroreductase (Figure 6). This can be said due to oxygen insensitive nitro-reductase have no effect of oxygen on its inhibition when incubated at shaking condition. Further confirmation of acetanilide as a metabolite indicated the involvement of N-acetyl transferase. An intermediate formation such as acetaminophen showed CYP450 mediated monooxygenase activation. The superoxide dismutase induction suggested the stress of super oxide generated during the process of biotransformation. The active form of enzymes in crude cytosolic fraction suggested that the bacterial strain is strongly involved in biotransformation producing necessary enzymes (Figure 7). The family of nitroreductase enzymes was observed in bacterial species and a lesser extent in eukaryotes useful in bioremediation, bio-catalysis, and chemotherapeutic treatment of tumor and ablation of specific cells [79]. Biotransformation of mono and dinitro-aromatic compounds was measured in sewage effluent under aerobic or anaerobic conditions by earlier researchers. Most of the nitrobenzene, nitro benzoic acids, nitrotoluenes, and dinitro-benzene have disappeared both in the presence and absence of oxygen [80]. A nonenzymatic reaction rarely leads a significant change in the structure of chemical compound [81]. It was reported that in presence of NADPH in cell extracts the 4-nitrobenzoate was transformed to 4-hydroxylaminobenzoate, 3,4 dihydroxybenzoate [82]. Antioxidant ways of defense in biological systems mainly consist of enzymatic and non-enzymatic reactions [83]. The formation of amines and catechol in increasing concentration during metabolism might have generated oxidative stress on the cell. To protect the DNA as well as plasma membrane form such oxidative damage super oxide dismutase, catalase, and peroxidase enzymes showed higher amplitude in experimental strain pseudomonas DL17 during this process. In case of non-polar nitro-aromatic compounds it had been stated that microorganisms can reduce the nitro groups but cannot cleave the aromatic ring [84]. In case of pseudomonas DL17 it was noticed that the higher activity of nitroreductase but little less than acetanilide hydroxylase and catechol 2, 3 dioxygenases (Figure 7). Thus as a concluding remark it could be stated that the key enzyme in bio-transformation might be nitro reductase leading to form p-phenylene diamine. The second step to acetanilide formation indicated that experimental bacteria used the defense machinery to protect it from the carcinogenicity supposed to be exerted by the new amine group formed on the aromatic ring. In a biodegradation report of nitrobenzene by pseudomonas pseudoalcaligenes JS45; the reduction of nitrobenzene proceeded through hydroxyl-amino benzene followed by rearrangement to 2 amino-phenol which then undergone meta cleavage. However the experimental bacterial strain pseudomonas DL17 showed ortho as well as meta cleavage [85]. The Figure 7 refers various enzymes induced and involved in biotransformation. The hike in the enzyme activity of catechol dioxygenase compared to the standard is supporting the ring fission of catechol might have followed ortho as well as meta cleavage pathway yielding cis-cis muconic acid and muconate semialdehyde [86].
It was noticed that the centrifugate showed pH 8.5 after experimental times although the broth used was buffered and adjusted at pH 9.0 in the beginning of experiment. This might be due to generation of organic acids such as muconic acid or else other. The dissolved alkali metals employed in the broth media could get exhausted. Bio-augmentation of dissolved alkali may minimize this problem as in other cases the biotechnologist does. However no other limitations observed using an alkaliphile pseudomonas DL17.
Conclusions
Biotransformation is an important process and a part of bio-degradation used in bioremediation. The study of such mineralization pathways explores various enzymes and sometimes gives novel metabolites for biotechnology or modern chemical science. From this study it is concluded that the toxicity could be reduced on bio-processing the industrial nitro-aromatic waste in alkaline habitat easily. Pseudomonas DL17 is a microorganism having indigenous ability to bio-transform the nitro-aromatic contaminants. The experimental microorganism might have gone through ortho and meta cleavage pathway initiating with nitro reduction of the contaminant and acetylation of successive metabolite generated. The enhanced acetanilide hydroxylase activity showed the Cyp 450 mediated monoxygenase had been played important role in polarization of aromatic ring and increasing the solubility of contaminant. The catalase and peroxidases are the enzymes able to transform a variety of compounds involved in free radical reactions. Such type of reactions yields oxidized or polymerized products to reduce the toxicity of contaminants. Super oxide dismutase induced might be engaged in minimizing oxidative stress generated. Extremophilic enzymes possess extreme stability, and the applications of these enzymes as biocatalysts are attractive because they are stable and active under such a condition which were previously regarded as incompatible with biological materials. Thus use of Alkaliphiles may be a good remedy to reduce environmental contamination and rejuvenate the natural ecosystem from alkali stable contaminants. The bacterial strain pseudomonas DL17 can be employed for bioremediation purpose at contaminated site as well as it could be a good source of various enzymes useful on commercial scale.